Both DNA barcoding and genome sequencing start with extracting DNA from a specimen. First the sample must be physically broken up to release molecules from the cells. This can be done by hand using a tiny pestle (usually for fresh samples) or in a machine (usually for dried specimens). Sand can be used to help grind the material, liquid nitrogen to freeze the material to make it more brittle, and chemical ‘buffers’ can help stop the DNA being broken during the process.
The DNA must then be precipitated out of the solution – which can be achieved by spinning at high speed in a centrifuge with an alcohol such as isopropanol – and washed. For some groups of organisms, such as plants, additional treatment may be needed to remove other molecules that might stick to the DNA. Commercial DNA extraction kits can also be used to simplify this process.
Electrophoresis – which separates the molecules in a solution by their size and electrical charge – is then used to check the quality and quantity of the DNA obtained.
Next-generation-sequencing (NGS) methods – which can read the sequence of many thousands of DNA pieces at once – have massively speeded up the sequencing of whole genomes. Several methods are available, including those created by Illumina, Oxford Nanopore and Pacific BioSciences.
Most NGS methods first require the creation of a ‘library’ of small fragments of DNA from the extracted sample, which together represent the whole genome of the organism. Each of the fragments in the library is treated (e.g., by the addition of small ‘adaptor’ sequences to its ends) to enable it to be ‘read’ by the sequencing machine.
For barcoding, only small, specific regions of the genome need to be sequenced. The region sequenced for barcoding will depend on the organism: for animals usually a region called COX1; for plants usually a combination of multiple regions.
Fresh barcode samples are sequenced using a process called ‘amplicon sequencing’, in which we amplify the specific barcode regions (amplicons) and then use NGS to sequence them. For the more fragmented DNA typically obtained from museum and herbarium samples, more specialised methods are needed. We are constantly improving our techniques to extract high-quality DNA, even from centuries-old specimens.
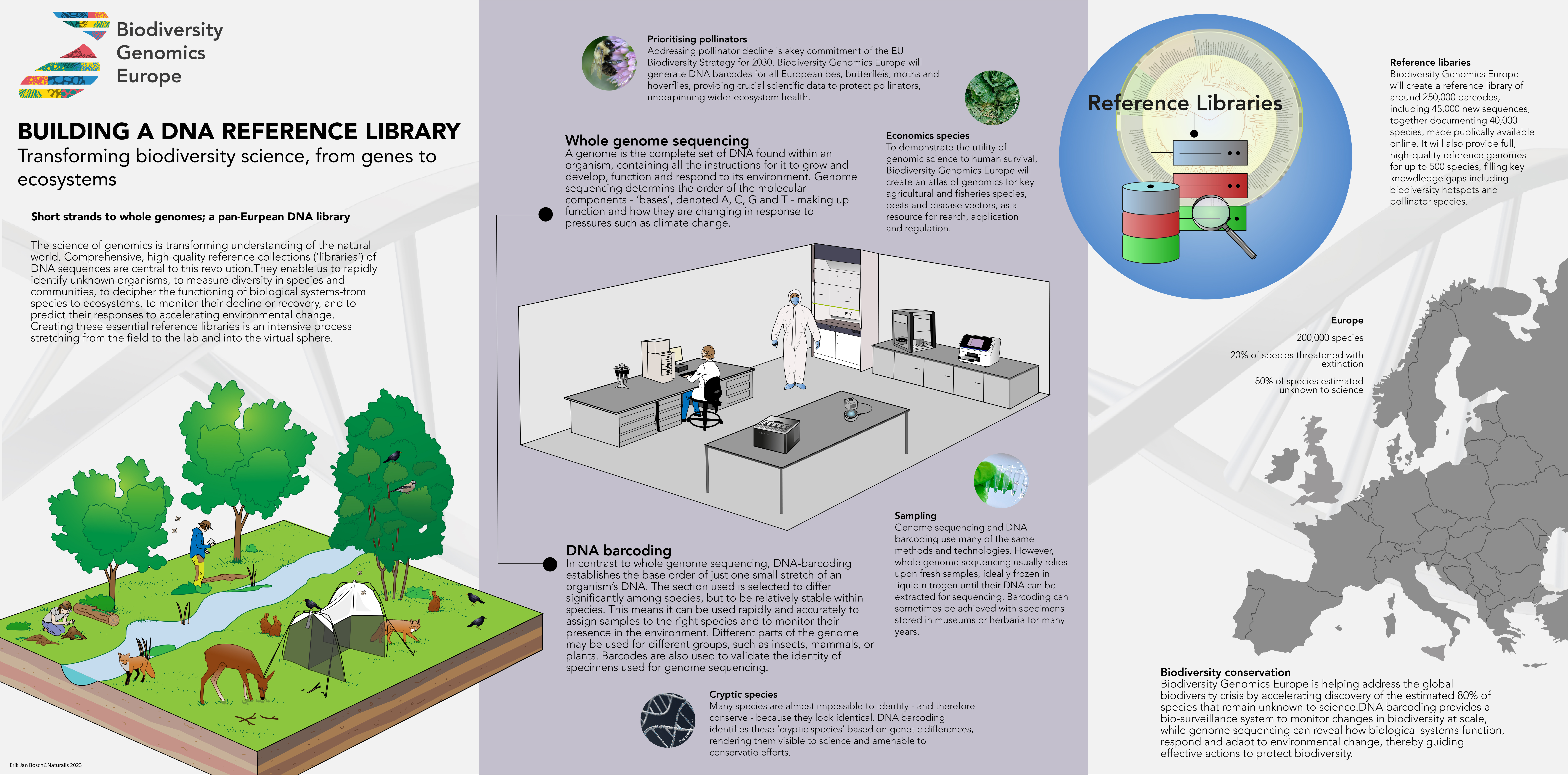
The infographic below shows the entire workflow of genomic research, from field sampling to application.